Tutorial
You have different option in using BAR 3.0: you can provide a sequence, either by accession or fasta file; GO Term, PFAM or PDB identifier, ligand code or a NCBI taxonomy code.
To try some possible searches, click on the suggested queries below the input box.
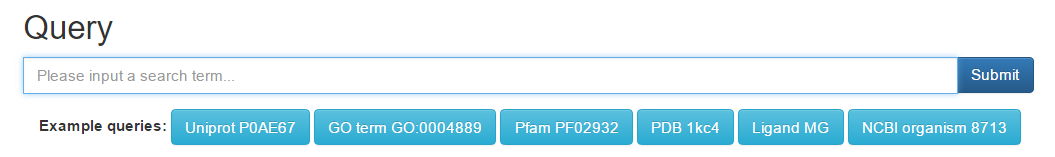
Option 1: type the accession of query sequence
Enter a UniprotKB accession P0AE67 in the query field and press Submit. This accession is in BAR 3.0, you will get a page with the cluster annotation.
Option 2: type the Fasta of query sequence
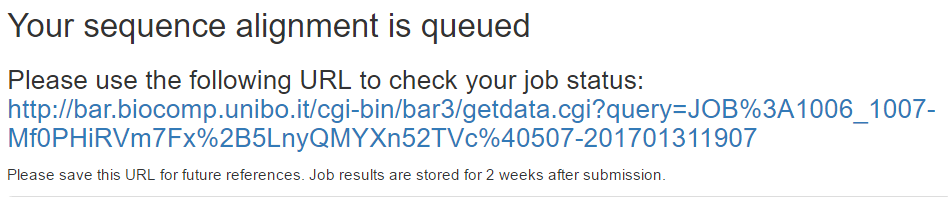
Enter a Fasta sequence in the bottom query field and press Submit. If the sequence is in BAR 3.0, you will get a page with the cluster annotation.
Otherwise, the sequence will be aligned to BAR 3.0 sequences and you will get a link to check the result.
Option 3: annotation search
Enter the GO term GO:0004889 in the query field and press Submit. The result will be a table listing all the BAR 3.0 clusters associated to the GO term after statistical validation.
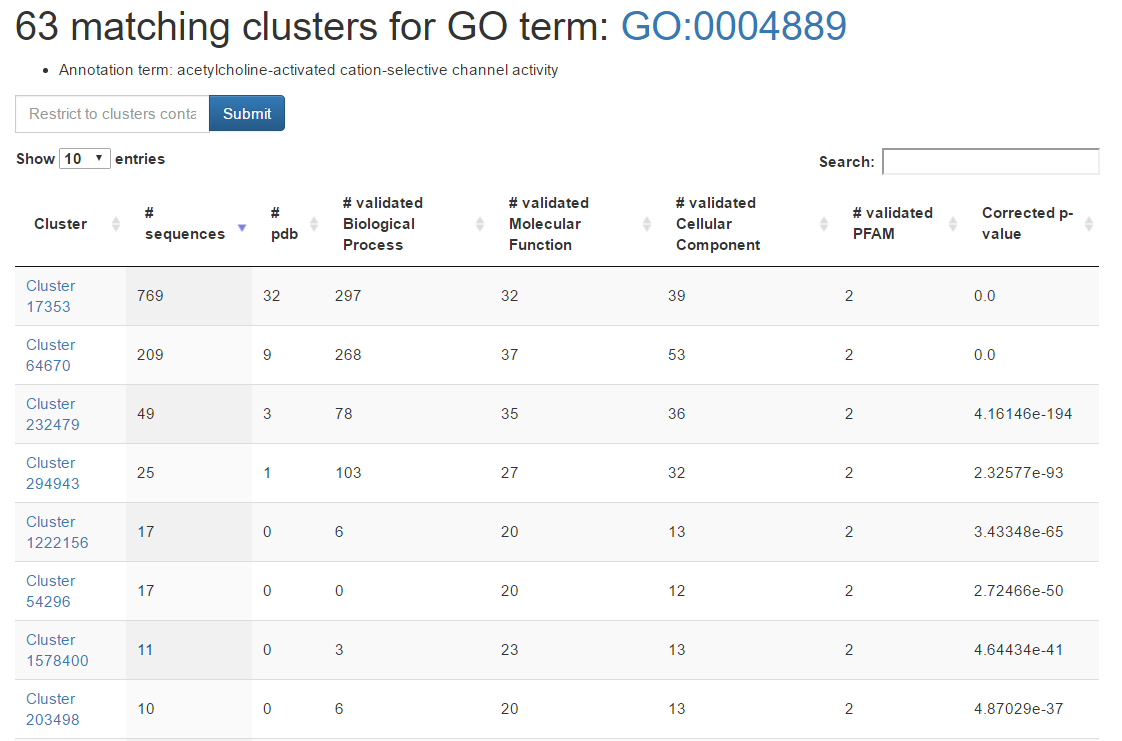
By clicking on the cluster number (first column) you will get a page with the cluster annotation.
The name of the GO term is displayed under the title. There is also the possibility to filter the results by specifying an NCBI Taxonomy identifier in the box on the top-left above the table. This filtering feature is available for GO term and PFAM queries only.
A similar table will be shown for queries by PFAM, PDB identifier, ligand, or NCBI taxonomy code in the query field.
Output
Cluster information
The cluster annotation page shows cluster statistics at the top:
- the number of sequences in the cluster, and how many from SwissProt
- the average lenght of the sequences in the cluster
- the number of different species of origin for the sequences, classified by domain
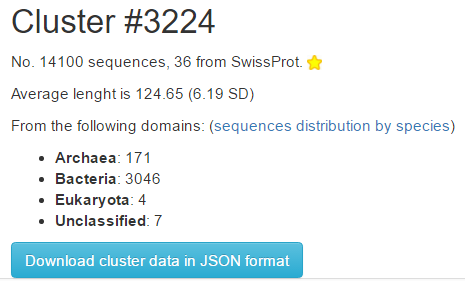
The button Download cluster data in JSON format allows to download all cluster information in a machine readable format. The schema for the JSON file is available here.
Matching sequence
The Query box shows information about the best match for the query sequence. This section reports the sequence identity (SI) and coverage (COV) of the best alignment against the cluster.

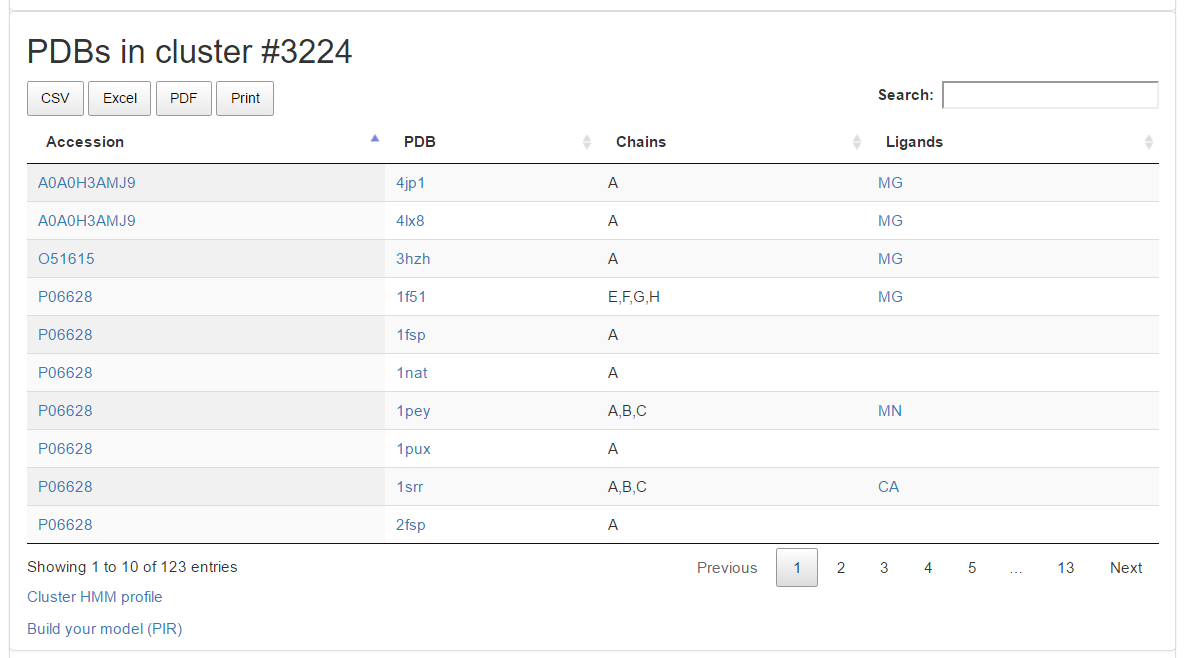
PDB in cluster
If the cluster contains structural information, the PDB section contains a table listing all the PDB chains associated to it, along with UniprotKB accessions and ligands. All rows links to the PDB files associated to sequences in the cluster. At the end of this section you can find links to a cluster HMM profile and a link to the downloadable alignment of the query sequence against it.
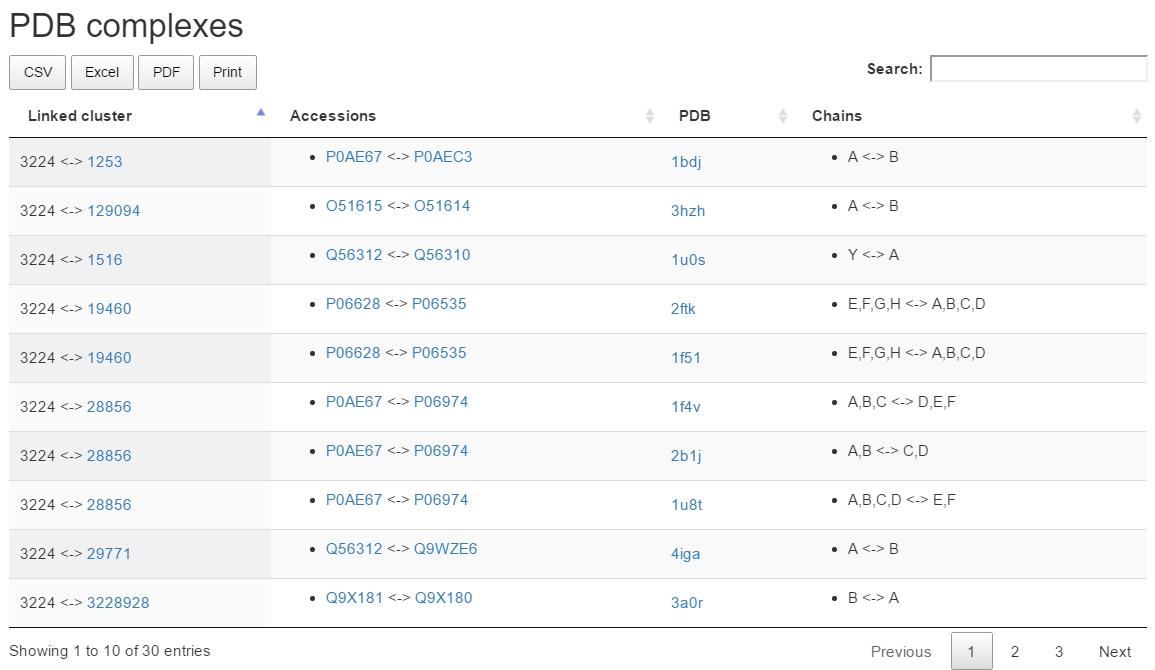
PDB complexes
The PDB Complexes section lists clusters containing sequences in complex with members of the result cluster. For each PDB complex, the table shows the linked cluster, the matching UniprotKB accessions, the PDB and its chains.
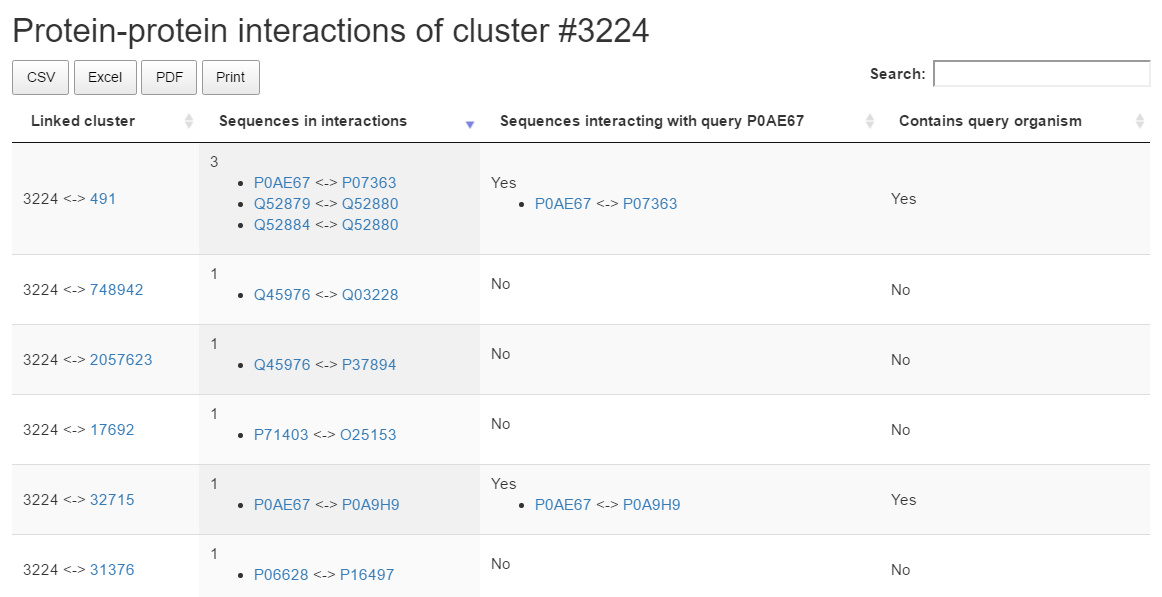
Protein protein iteraction
The Protein-protein interactions section lists clusters containing sequences associated in IntAct to members of the result cluster. For each matching cluster the table displays the number of interacting pairs, if the query sequence is in iteraction and with whom, and if the matching cluster contains sequences from the same organism as the query sequence.
GO terms
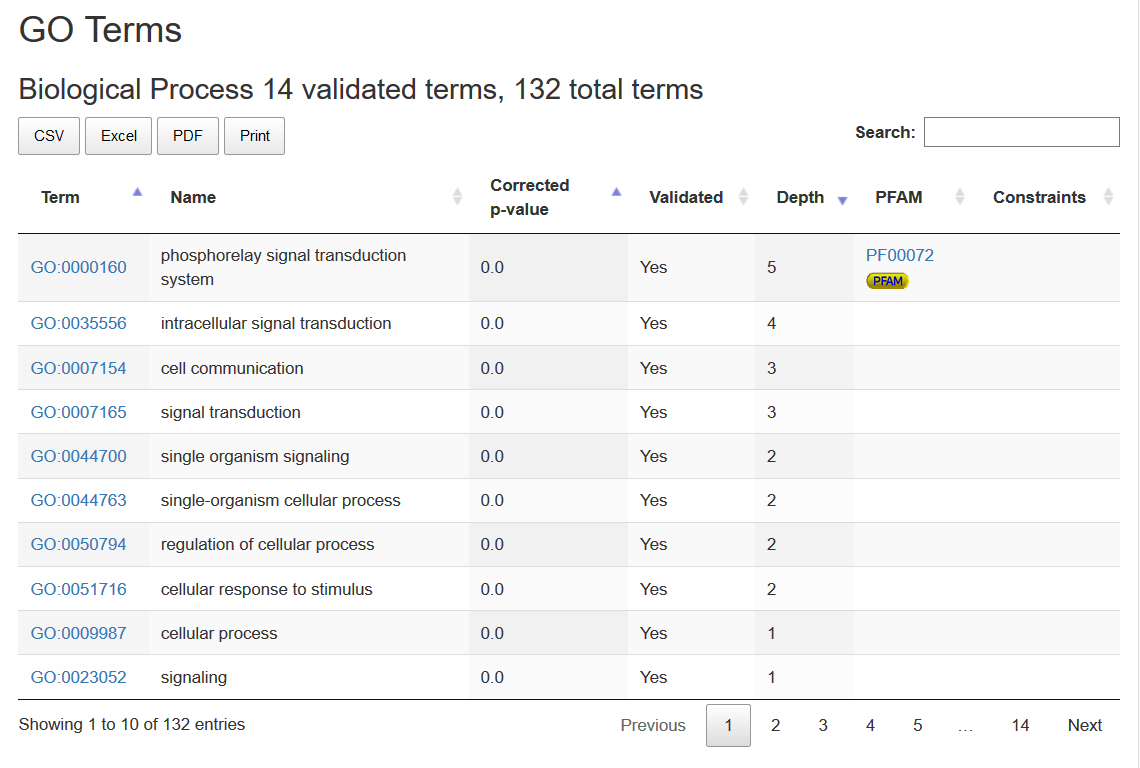
Next there are GO Term and PFAM annotations are listed. The heading of each annotation type (Biological Process, Molecular Function, etc) reports the number of statistically validated terms over the total number of annotation terms of the same type in the cluster.
Each term links to the relative entry in its database of origin. Statistically validated terms are marked as "yes" in the Validated column. Terms are sorted by P-value, from the lowest to the highest, and by term depth in the ontology (from deeper to shallower).
Statistically validated PFAM may bring associated GO terms to the validated status. Such GO terms are marked by an icon into the PFAM column, that also contains the associated PFAM.
If the GO term may be associated only to sequences from a specific taxonomy, or in case it should never be associated to sequences from a specific taxonomy, such limitations are shown into the Constraints column.
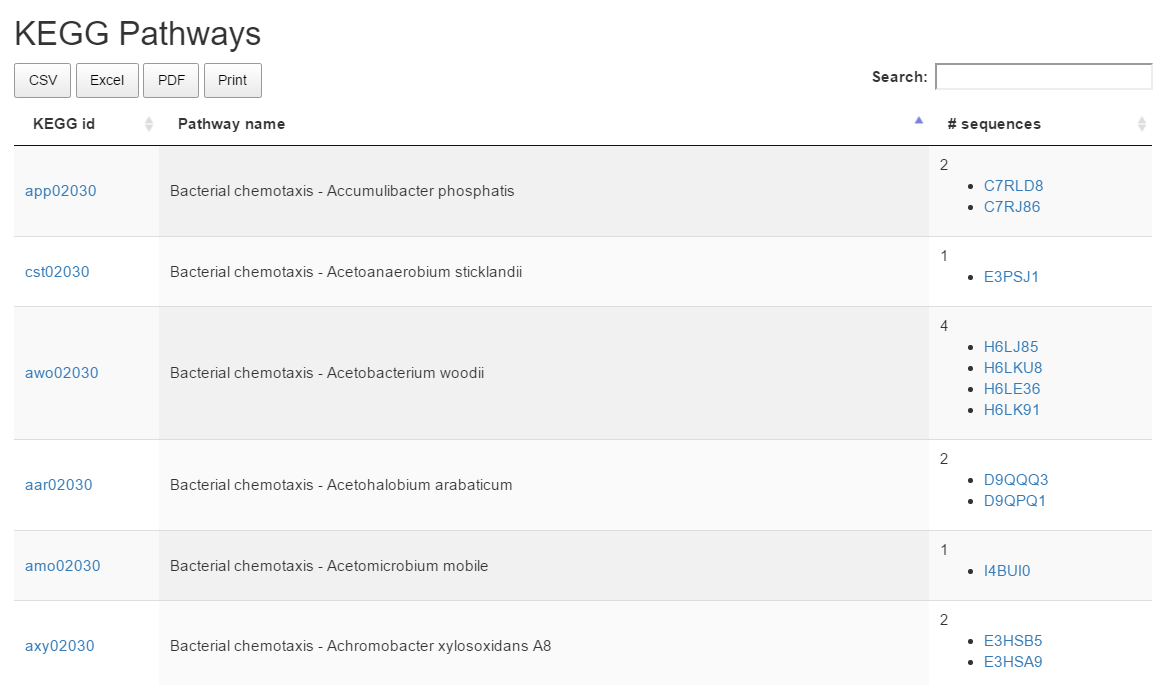
KEGG pathways
The KEGG Pathways section lists KEGG pathways associated to sequences form the result cluster.