Tutorial
Example 1: retrieve the statistically validated SUS-BAR annotation of a UniProtKB or Ensembl protein.
For any pig protein sequence of the UniProtKB or Ensembl database you can check which annotation can be retrieved from the SUS-BAR database. In this case the search can be done selecting the keys "UniProtKB accession" or "Ensembl accession" and entering the protein UniProtKB or Ensembl code.
The retrieved result is a table reporting the name of the protein, its length and the corresponding SUS-BAR cluster (when available). As an example you may consider searching the UniProtKB protein O02837 and retrieving the following table:
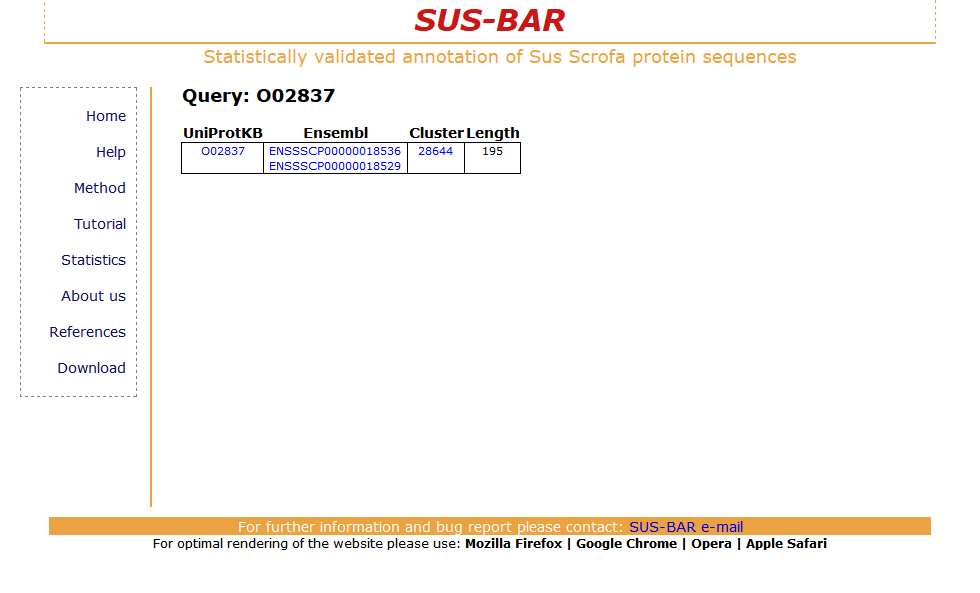
The UniProtKB protein O02837 has two counterparts in the Ensembl database with the accession codes ENSSSCP00000018529 and ENSSSCP00000018536 (reported in the table where the codes are linked to the corresponding data bases). The sequence with a length of 195 residues falls into cluster number 28644. To obtain the list of all the cluster specific annotations that th protein can inherit from the cluster click on the cluster number. The new page displays the cluster features, a list of the PDB structures that are templates for the cluster sequences, Pfam statistically validated domains and the list of the statistically validated GO terms (Biological process, Cellular Component and Molecular function) that are cluster specific. The list of the protein sequences in the cluster can be also downloaded.
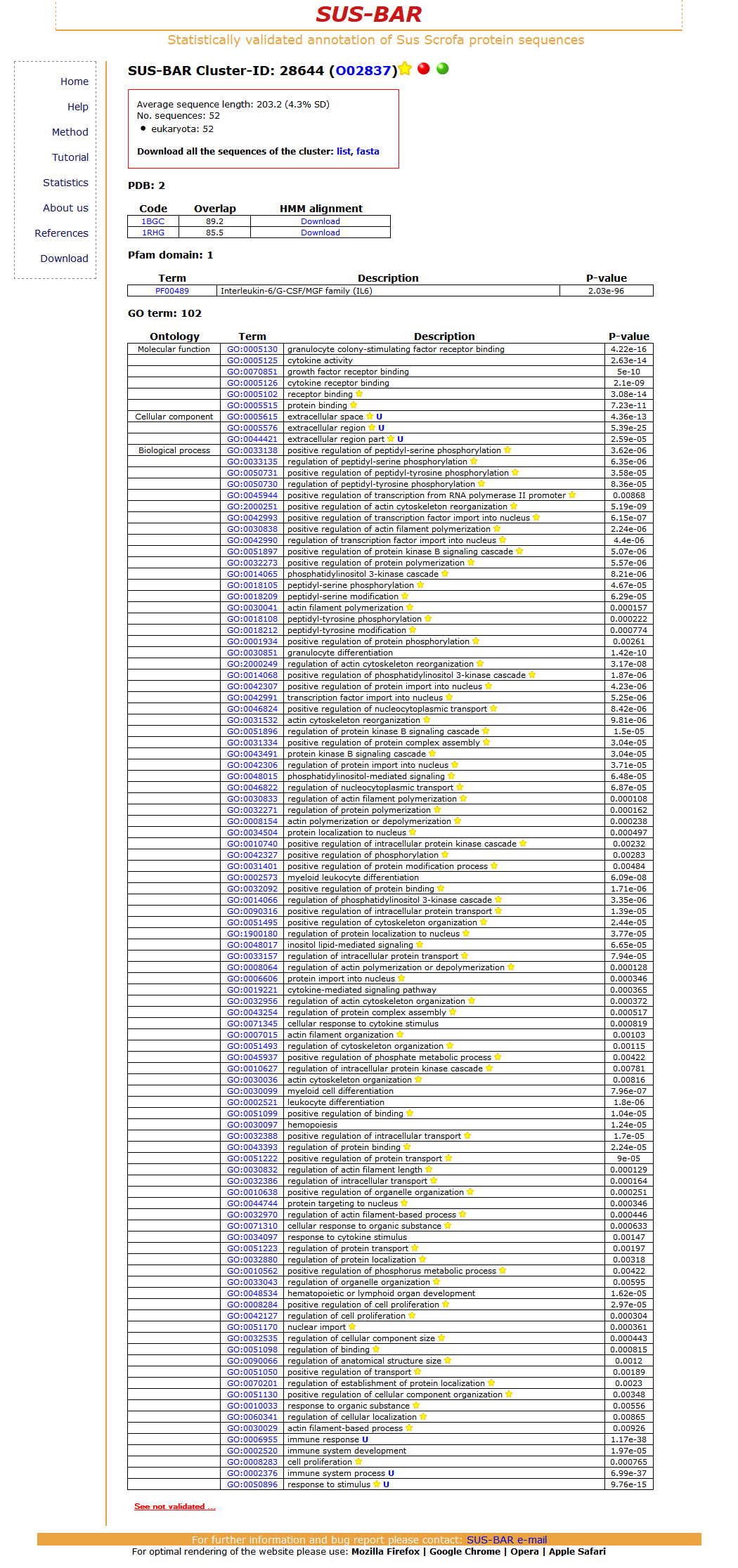
In this cluster two PDB templates (1BGC and 1RHG) are present with an overlap of 89.2 and 85.5 % with the corresponding sequences in UniProtKB are present. These structures can be used to model the pig protein sequence O02837 and the target to the template alignment is provided by a cluster specific HMM that encodes the multiple sequence alignment of all the sequences in the cluster [see Method]. These alignments are downloadable. The pig protein sequence O02837 by falling into this cluster inherits the statistically validated (P-value) Pfam domain (PF00489) that in this case corresponds to the Interleukin-6/G-CSF/MGF family (IL6) domain. 25 statistically validated GO terms of the three main routes (Biological Process, Molecular Function, and Cellular Component) are cluster specific and are inherited by the pig protein. The statistically validated Go terms are listed and sorted by their P-value (on the top terms with the lowest P-value). All the other GO and Pfam terms that are in the cluster (if present) but are not validated by our procedure can be also downloaded. In our annotation system the pig protein is therefore enriched with a template for its modeling and GO terms that are statistically validated as compared to the annotation in the corresponding UniProtKB file.
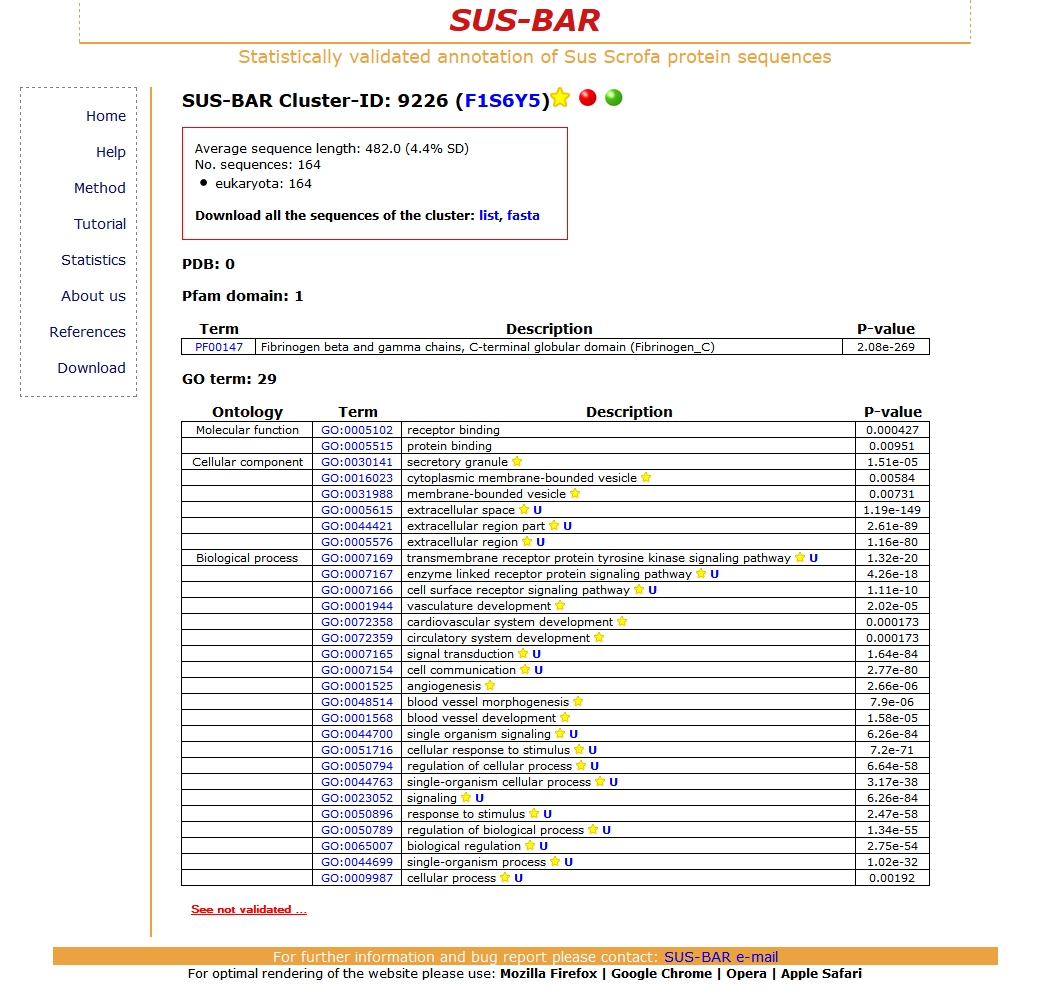
A second search is relative to the UniProtKB pig protein F1S6Y5. This protein is predicted and poorly annotated in the UniProtKB data base. In SUS-BAR by falling into Cluster 9226 it receives 14 statistically validated GO terms not associated before to the sequence. Furthermore the Pfam term PF00147-Fibrinogen beta and gamma chains, C-terminal globular domain (Fibrinogen_C), associated to the protein also in its UniProtKB file, and is statistically validated in SUS-BAR.
Example 2: search all the pig protein sequences annotated with a specific annotation (PDB, Pfam and GO) in the SUS-BAR clusters.
Suppose you are interested in all pig proteins involved in the process of smell. In the home page of the SUS-BAR database you can just search the word "smell" selecting "SUS-BAR GO term" in the drop-down menu on the right of the search field. The result is a table that lists all the pig proteins that fall into clusters where is present a specific statistically validated GO term that contains the word "SMELL" in its definition.
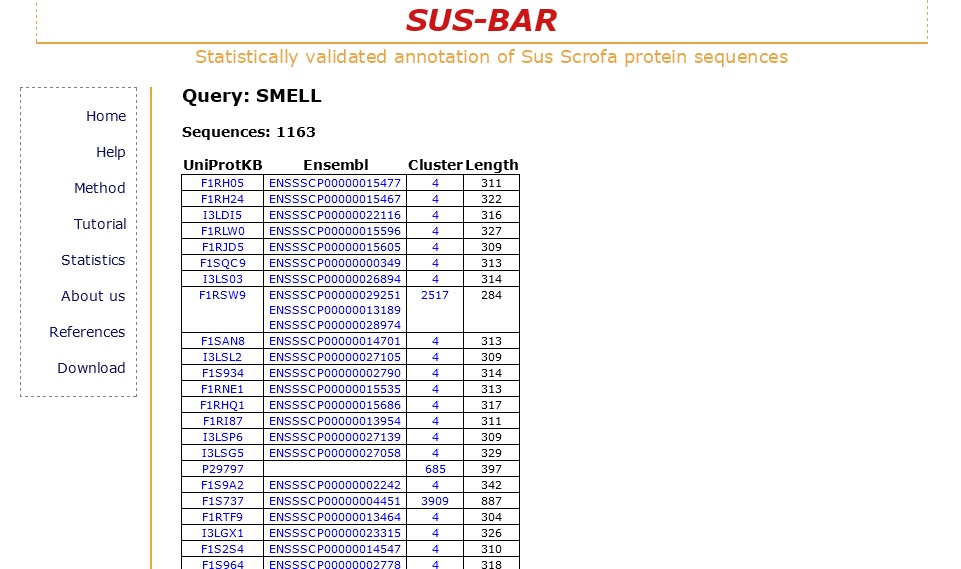
For each sequence its accession code, the cluster where the sequence is found and its length are listed. The list is downloadable.
Note that if you are interested in a particular GO term you can first check in the Gene Ontology web server how many different terms contain the word "smell" in their definition. In the SUS-BAR database to avoid errors you can search directly the accession code or the entire definition of a term, in this case: "GO:0007608" or "sensory perception of smell".
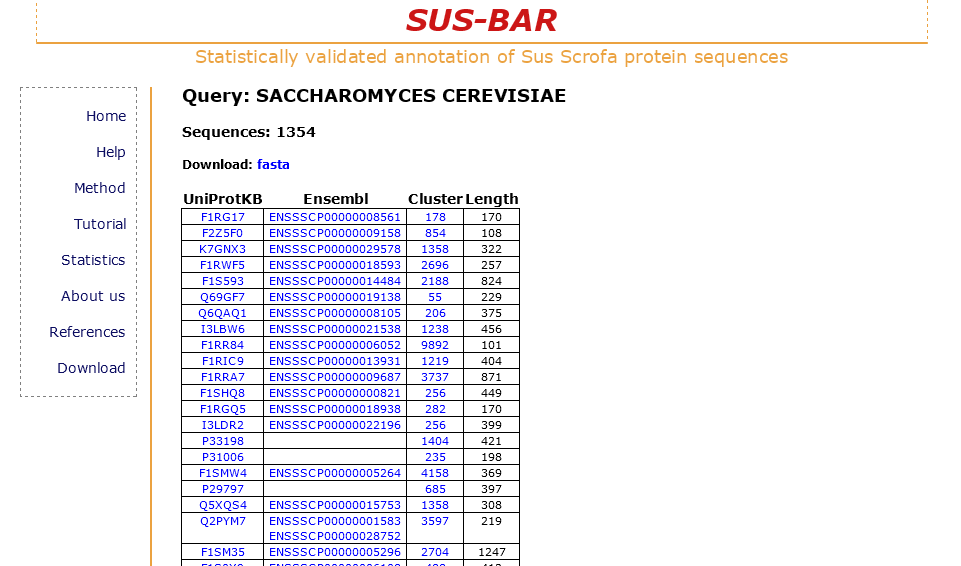
Example 3: search all the pig protein sequences falling into clusters containing sequences from other specific organisms or taxa.
With the "SUS-BAR Organism" option you have the opportunity to retrieve all the clusters that contain pig protein sequences together with those of other specific organisms. The result page shows the same information as seen in the previous examples. Below you find the result table displayed when “Saccharomyces cerevisiae” is searched. In this case a total of 1,798 pig protein sequences are retrieved and can be downloaded and analyzed.
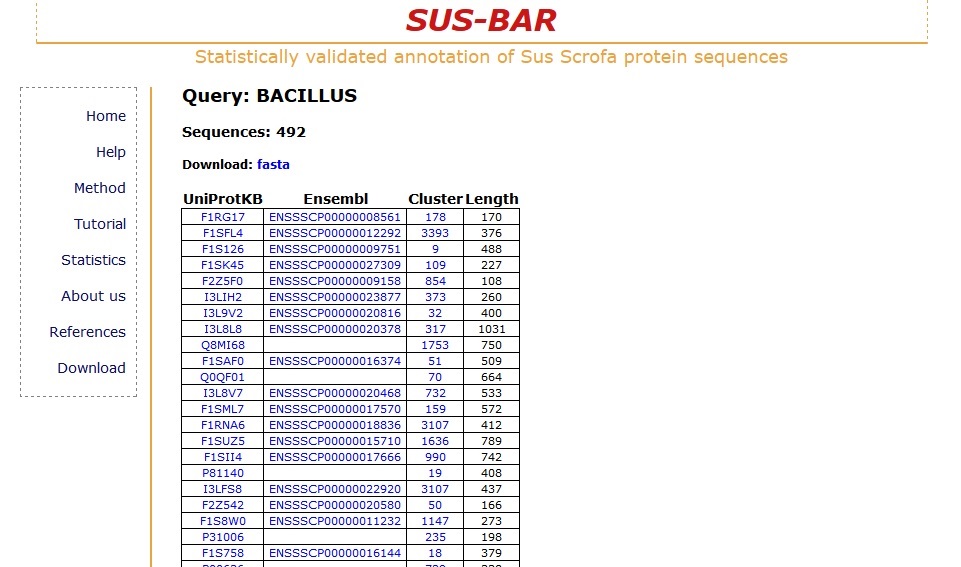
With the "SUS-BAR taxon" option you have the opportunity to retrieve all the clusters that contain pig protein sequences together with those of other specific taxa. In particular suppose you want to search all the pig sequences that coexist in clusters where there are some organisms of the "bacillus" genus. The search returns all the pig sequences that fall into clusters where there protein sequences from organisms that contain the word "bacillus" in their lineage.